Abstract
Background: Dysregulation of histone modifying marks and their modulators leads to aberrant gene expression and could contribute to leukemogenesis via misregulation of gene transcription of tumor suppressor genes and oncogenes. Although understanding of the role of the epigenome in cancer has expanded greatly, until recently there has not been a comprehensive characterization of protein expression patterns for multiple histone modifications, histone modification-related proteins (HistModProt), or their association with clinical characteristics in AML. We recently demonstrated that adult AML HistModProt form recurrent patterns of expression that predict prognosis (van Dijk et al. 2018). We now seek to determine whether HistModProt expression can predict prognosis in pediatric AML.
Methods: We simultaneously analyzed expression of 7 histone modification marks (H3 core protein, H3K4me2 (2 antibodies), H3K4me3, H3K9me2, H3K27me3, H3K36me3) and 17 HistModProt (ASH2L, BMI1, BRD4, NCL, CLPP, HDAC1, HDAC2, HDAC3, HDAC6, hnRNPK, JMJD6, KDM1A, NPM1 (2 antibodies), SIRT1, SIRT6, WTAP) in 505 de novo pediatric AML patients by the reverse phase protein array (RPPA) methodology. Expression was compared to 20 non-malignant CD34+ bone marrow derived samples. Patients were clustered by the progeny clustering algorithm coupled with k-means, which computationally calculated the optimal number of clusters. Patients were treated according the COG Phase 3 AAML1031 trial with a 1:1 randomization to cytarabine (ara-C), daunorubicin, etoposide (ADE) ± the proteasome inhibitor bortezomib.
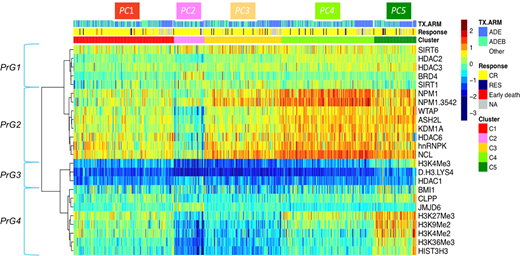
Results: Cluster analysis identified 4 groups of correlated Protein Groups (PrG) that defined 5 recurrent Patient Clusters (PC, figure 1). The first PrG consisting of HDAC's, SIRT's and BRD4 showed homogeneous normal range expression across all patients. PrG2 contains most varying expression of HistModProt and was therefore termed as modulators. PrG3, miscellaneous defined (misc) group, had universally low expression across all patients. PrG4 consists of the histone marks including total H3. Interestingly, all variation between the PC was due to modulation within PrG2 modulators and PrG4 histone marks.
Patient protein patterns in PC1 were identified as closest to that of the normal bone marrow derived CD34+ cells by performing linear discriminant analysis and were therefore defined as most normal-like cluster. PC2 had very low expression of both PrG2 and PrG4, representing a more deactivated/off cluster. PC3 had somewhat low histone marks, with higher modulators relative to the CD34+ normal-like PC1. PC4 showed relative normal levels of histone marks but had the highest levels of modulators (i.e. NPM1, NCL and hnRNPK). PC5 had elevated levels of both modulators and histone marks and therefore PC4 and PC5 were considered to represent a more activated/on protein signature that was associated with a higher proliferative potential, as manifested by higher WBC, and percent peripheral blasts and absolute blood count (all p < 0.001). These findings correlate with clinical features of adult AML patients with upregulated HistModProt. However, in adults, different proteins (e.g. high BRD4 and KDM1) were more prominent in the upregulated HistModProt signature. HistModProt PC membership did not correlate with outcome overall (OS) or event free survival (EFS). However, pediatric patients with the on signature (PC4 and PC5) that received the ADE + bortezomib regimen had a trend towards longer EFS compared to those treated with ADE (P = 0.055), whereas PC2 patients did not benefit from the addition of bortezomib (p = 0.43).
Conclusion: Similar to adult AML patients, recurrent patterns of HistModProt were observed in pediatric AML and associates with outcome. Relative low expression of histone marks and HistModProt correlated with favorable outcomes in both adults and pediatrics suggesting that these patients contain a more open chromatin state with potential for diagnostic and prognostic implications in AML at all age.
Figure legend: RPPA-based heatmap of 505 de novo pediatric AML samples containing 7 histone marks and 17 HistModProt. The five cluster colors along the top bar delineate the five identified protein clusters (PC). Protein expression is normalized for each protein to range from the lowest (blue) to the highest (red) compared to non-malignant CD34+ cells. Identification of protein groups (PrG's) is shown.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal